Statistical Analysis
| Total Publications: | 383 |
| Total Individuals: | 2182 |
| Total Genes: | 8058 |
| Total Variants: | 34,689 |
Download all entries from MosaicBase 
1. Publications
1. Publications
383 published papers were manually reviewed from 1989 to June 2018 and the genomic and phenotypic annotations of each mosaic variant were curated and integrated into the database. Starting from the recognition of the important role of genomic mosaicism in genetics, studies on genomic mosaicism burgeons and the number of publications increases steadily through the past decades.
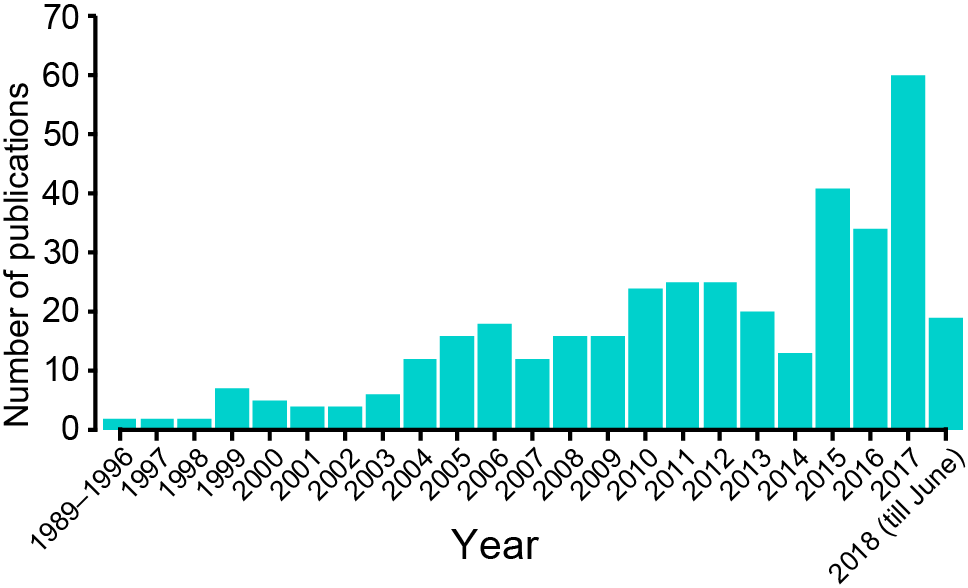
Figure 1. Distribution of publication date of papers collected in this database.
2. Individuals
In summary, 2182 individuals carrying postzygotic mosaic variants are included in the database. There are mainly two kinds of mosaic variants in our collection: the mosaic variants related to non-cancer diseases and those detected in 422 healthy individuals. Disease-related mosaicism includes the transmission of mosaic variants from parents or grandparents to offspring. The field 'whose mosaic' is denoted as 'grandparents'/'parents' in this situation. Disease-related mosaicism also include the event that the mosaic variants in one dividual directly lead to the abnormal phenotype or disease. The field 'whose mosaic' is denoted as 'patient' in this situation. The number of individuals in each categories are shown below.
|
|
|
Table 1. Number of individuals in this database. According to the description of phenotype in published papers, phenotype of each individual is classified into one of the three categories including: (1) asymptomatic, (2) milder phenotype that fulfills some but not all the disgostic criteria, and (3) all the diagnostic criteria of the specific disease are fulfilled. Fractions of individuals in each category are shown in the pie chart. The moasic allele frequency (MAF) information estimated in the original publication was also integrated into the database if available. The percentage of individuals with mosaic allele frequency in each decile is presented here.
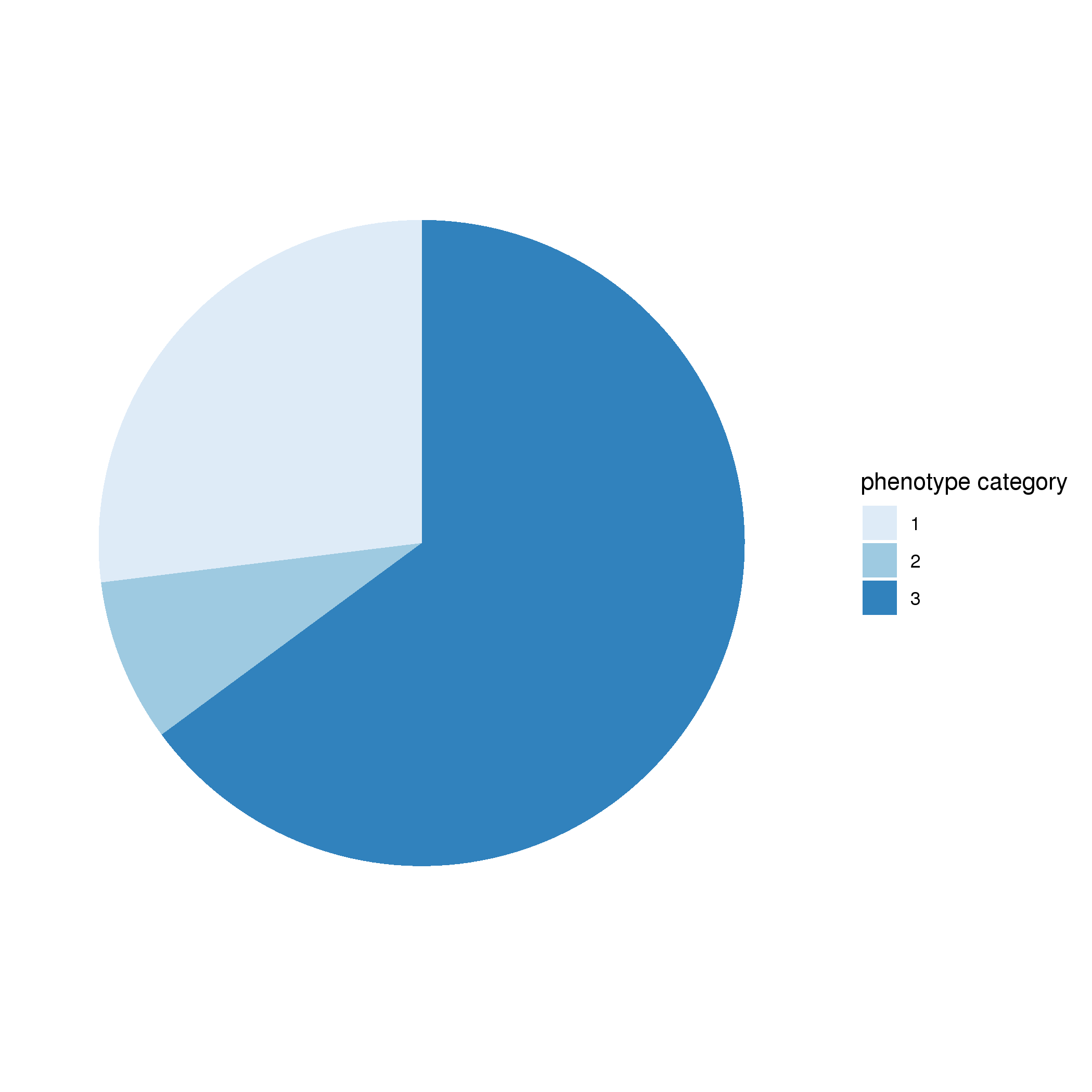
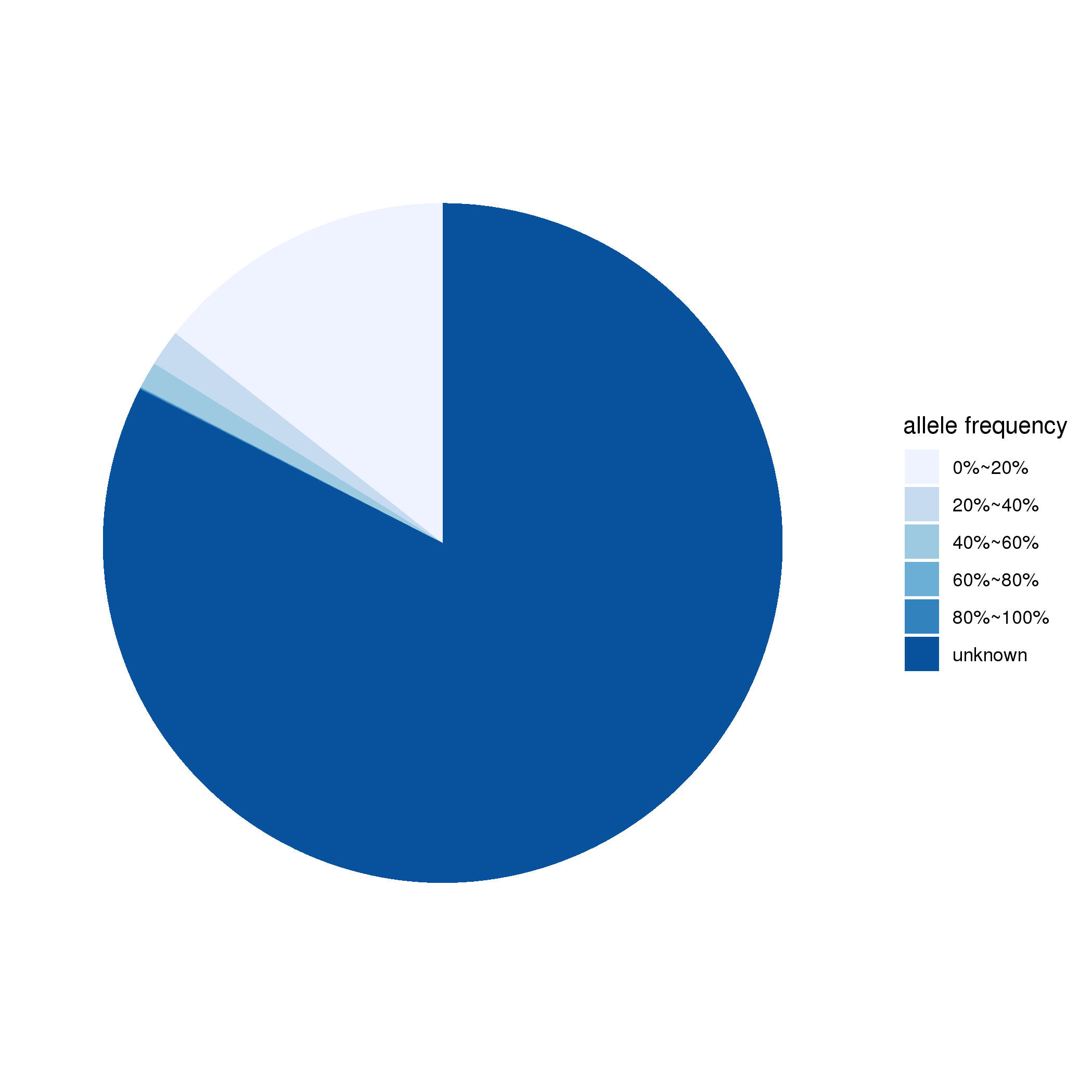
Figure 2. (Left) Percentage of individuals in each phenotype category; (Right) Percentage of mosaic variants in each decimal MAF.
3. Landscape of postzygotic single-nucleotide and small indel variants.
So far, 34689 postzygotic mosaic variants from non-cancer individuals are collected and integrated into this database. The genomic distribution and mutational spectrum of these mosaic muations are shown here.
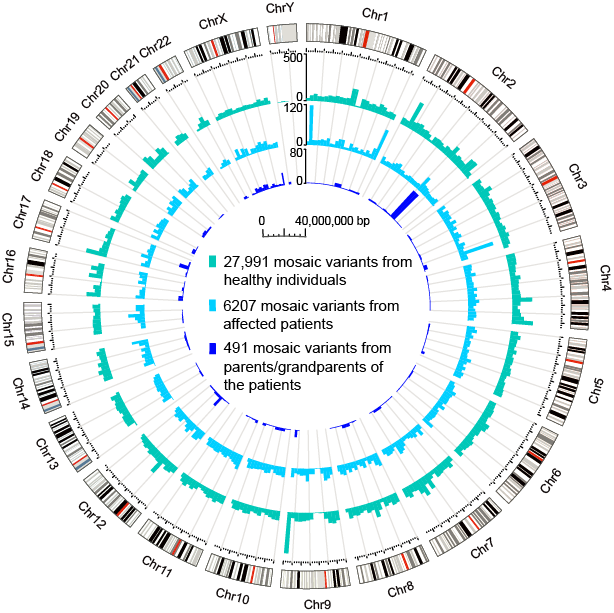
Figure 3. Genomic distribution of mosaic variants collected in this database. Circos plot of the mosaic variants collected in MosaicBase. Histograms demonstrate the number of variants for each 1Mb genomic window. Chromosomal bands are illustrated in the outer circle with centromeres colored in red.
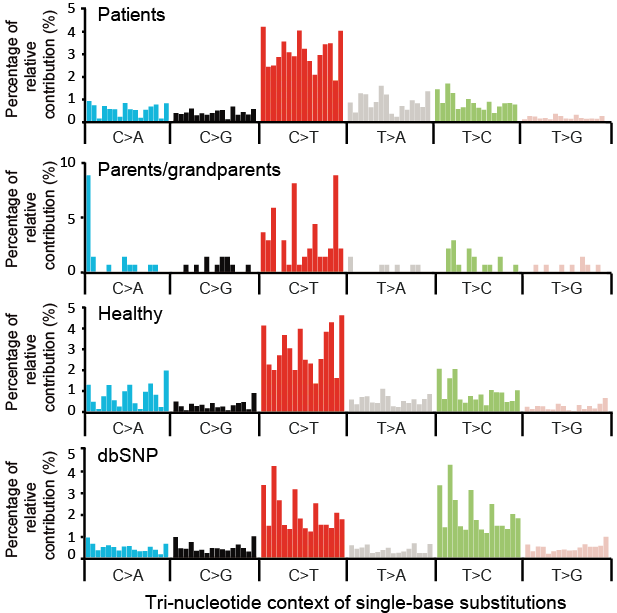
Figure 4. Statistical analysis of base substitution spectrum of mosaic SNVs in MosaicBase. Tri-nucleotide mutation spectrum of mosaic and inherited SNVs. Common variants with population allele frequency higher than 10% in dbSNP (version 137) are also shown. The description of tri-nucleotide mutation spectrum can be found here
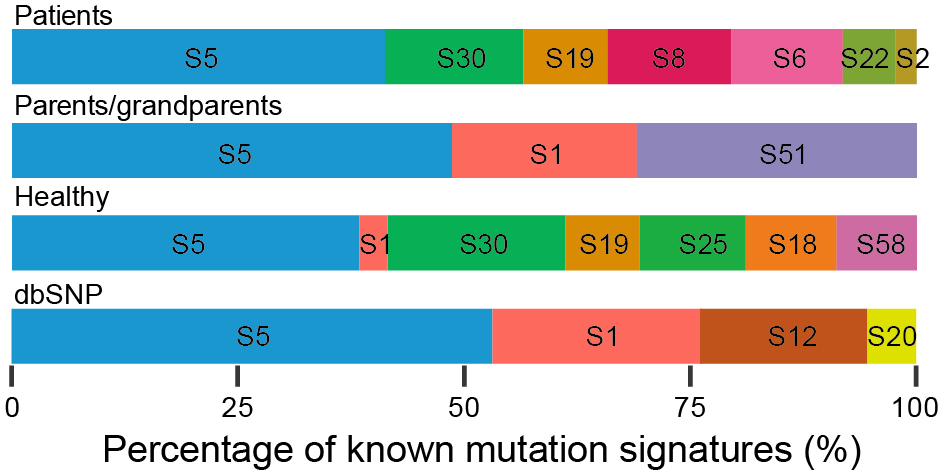
Figure 5. Proportion of single base mutation signatures for mosaic and inherited SNVs. Signatures are decomposed to reach a prediction rate of >96%. Common variants with population allele frequency higher than 10% is also shown and compared.
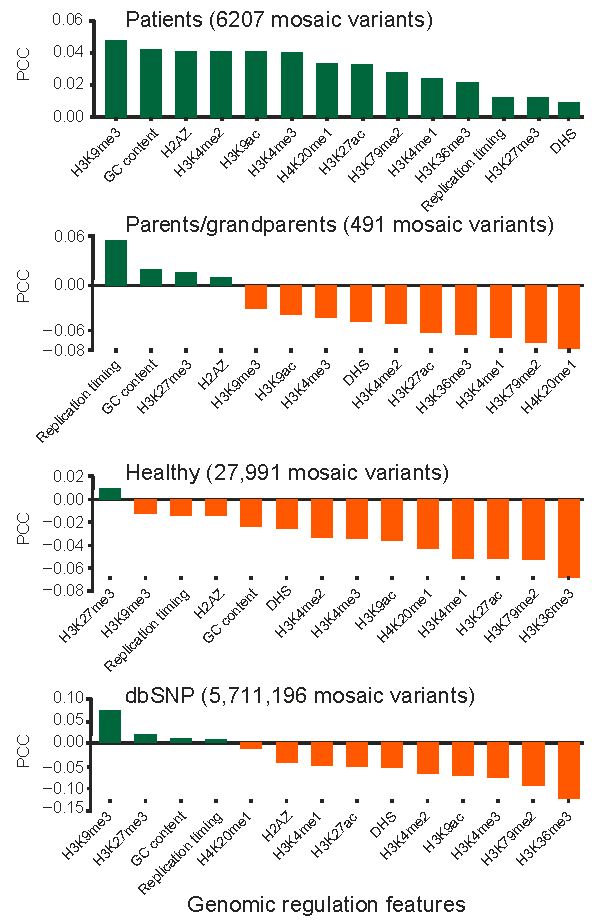
Figure 6. Correlation of the density of mosaic variants and known genomic regulation features. GC contents, Dnase I hyper sensitive postions, Replication timing, Histone modification profiles from GM12878 are compared. Y axis is the genome-wide Pearson correlation coefficient with a window size of 1MB. Variants collected in MosaicBase are compared with common SNPs in dbSNP (version 137) with population AF > 10%. H2AZ, Histone 2A.Z variant; DHS, Dnase I hyper sensitive; PCC, Pearson correlation coefficient.
4. Mutant allelic fraction (MAF) and carrier phenotype.
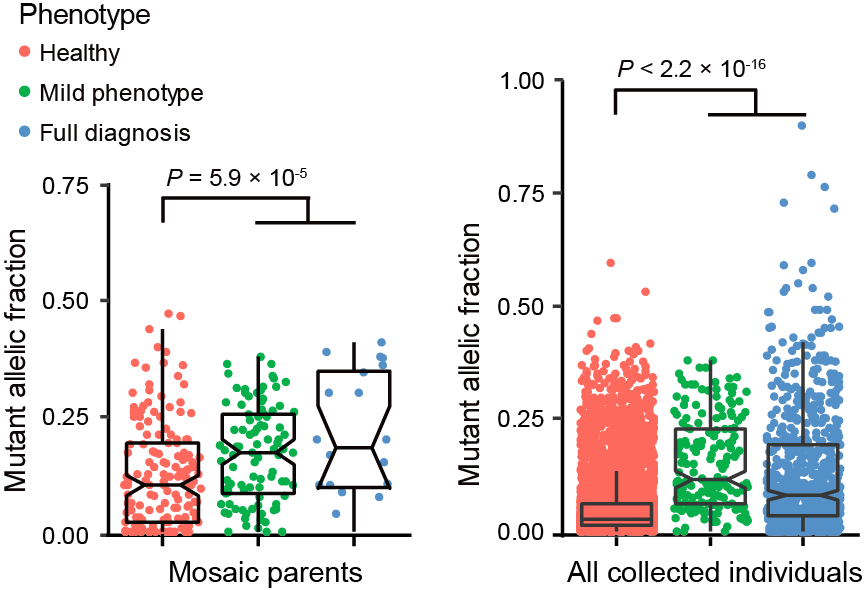
Figure 7. The general relationship between MAF of mosaic variant and carrier phenotype. No significant difference was found when we considered the variants identified from all individuals. However, when we went into mosaic variants in "parent" group, the mosaic variants in parents with milder or full disease phenotype had significantly higher MAF than those in asymptomatic parents.